Harvard Researchers Could Store the New York Public Library in a Teaspoon of Protein
Researchers at Harvard University seem to have devised a way that could fit library-sized data into a teaspoon of protein.
“Think storing the contents of the New York Public Library with a teaspoon of protein,” says Brian Cafferty, Ph.D., first author on the paper that describes the new technique and a postdoctoral fellow in the lab of George Whitesides, Ph.D., a Core Faculty member of Harvard’s Wyss Institute for Biologically Inspired Engineering, and the Woodford L. and Ann A. Flowers University Professor at Harvard University.
The work was performed by Brian Cafferty, Ph.D., first author on the paper that describes the new technique and a postdoctoral fellow in the lab of George Whitesides, Ph.D., a Core Faculty member of Harvard’s Wyss Institute for Biologically Inspired Engineering, and the Woodford L. and Ann A. Flowers University Professor at Harvard University. The researchers also worked with Milan Mrksich, Ph.D., and his group at Northwestern University.
“At least at this stage, we do not see this method competing with existing methods of data storage,” Cafferty says. “We instead see it as complementary to those technologies and, as an initial objective, well suited for long-term archival data storage.”
Cafferty’s chemical tool might not replace the cloud. But the filing system offers an enticing alternative to biological storage tools like DNA. Recently, scientists discovered how to manipulate our loyal guardian of genetic information to encode more than just eye color. Researchers can now synthesize DNA strands to record any information, including cat videos, diet trends, and cooking tutorials (whether they should is another question).
But while DNA is small compared to computer chips, the macromolecule is large in the molecular world. And, DNA synthesis requires skilled and often repetitive labor. If each message needs to be designed from scratch, macromolecule storage could become long and expensive work.
“We set out to explore a strategy that does not borrow directly from biology,” Cafferty says. “We instead relied on techniques common in organic and analytical chemistry, and developed an approach that uses small, low molecular weight molecules to encode information.”
With just one synthesis, the team can produce enough small molecules to encode multiple cat videos at a time, making this approach less labor intensive and cheaper than one based on DNA. For their low-weight molecules, the team selected oligopeptides (two or more peptides bonded together), which are common, stable, and smaller than DNA, RNA or proteins.
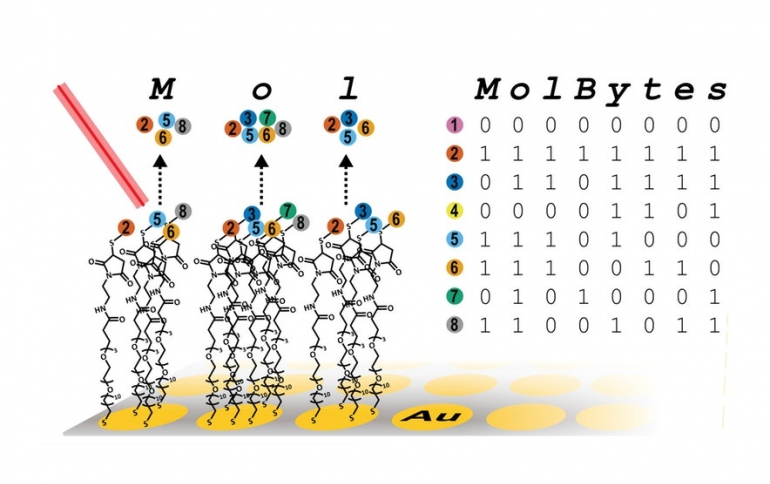
Oligopeptides also vary in mass, depending on their number and type of amino acids. Mixed together, they are distinguishable from one another, like letters in alphabet soup.
Making words from the letters is a bit complicated: In a microwell—like a miniature version of a whack-a-mole but with 384 mole holes—each well contains oligopeptides with varying masses. Just as ink is absorbed on a page, the oligopeptide mixtures are then assembled on a metal surface where they are stored. If the team wants to read back what they “wrote,” they take a look at one of the wells through a mass spectrometer, which sorts the molecules by mass. This tells them which oligopeptides are present or absent: Their mass gives them away.
Then, to translate the jumble of molecules into letters and words, they borrowed the binary code. An “M,” for example, uses four of eight possible oligopeptides, each with a different mass. The four floating in the well receive a “1,” while the missing four receive a “0.” The molecular-binary code points to a corresponding letter or, if the information is an image, a corresponding pixel.
With this method, a mixture of eight oligopeptides can store one byte of information; 32 can store four bytes; and more could store even more.
So far, Cafferty and his team “wrote,” stored, and “read” physicist Richard Feynman’s famous lecture “There is plenty of room at the bottom,” a photo of Claude Shannon (known as the father of information theory), and Hokusai’s woodblock painting The Great Wave off Kanagawa. Since the global digital archive is estimated to hit 44 trillion gigabytes by 2020 (ten times that of 2013), an image of a tsunami seems appropriate.
Right now, the team can retrieve their stored masterpieces with 99.9% accuracy. Their “writing” averages 8 bits per second and “reading” averages 20 bits per second. Although their “writing” speed far outpaces writing with synthetic DNA, reading could be both quicker and cheaper with the macromolecule.
But, with faster technology, the team’s speeds are sure to increase. An inkjet printer, for example, could generate drops at rates of 1,000 per second and cram more information into smaller areas. And, improved mass spectrometers could take in even more information at a time.
The team could also improve the stability, price, and capacity of their molecular storage with different classes of molecules. Their oligopeptides are custom-made and, therefore, more expensive. But future library builders could purchase inexpensive molecules (like alkanethiols) that would cost just one cent to record 100,000,000 bits of information.
Unlike other molecular information storage systems, which rely on one specific molecule, this approach can use any malleable molecule as long as it can be manipulated into distinguishable bits.
Oligopeptides—and similar choices—are already resilient. “Oligopeptides have stabilities of hundreds or thousands of years under suitable conditions,” according to the paper. The hardy molecules could endure without light or oxygen, in high heat and drought. And, unlike the cloud, which hackers can access from their favorite easy chair, the molecular storage can only be accessed in person. Even if a thief finds the data stash, a little chemistry is needed to retrieve the code.